Dive deeper into the universe of the Ruminant Holobiont with our newly unveiled infographic, specially crafted for the HoloRuminant glossary, where science meets visual storytelling!
ALGORITHM: A set of detailed, step-by-step instructions or rules to be followed by a computer.
ALLELE: One of two or more forms of a gene or a genetic region. A population usually includes multiple alleles at specific locations of the DNA/genome.
ANAEROBIC FUNGI: A group of microorganisms that grow without oxygen; these degrade plant fibre.
ARCHAEA: Microorganisms without an organised nucleus (prokaryotes) that can be found in the rumen; they are the sole methane producers in the rumen and most often do this by feeding on hydrogen and carbon dioxide.
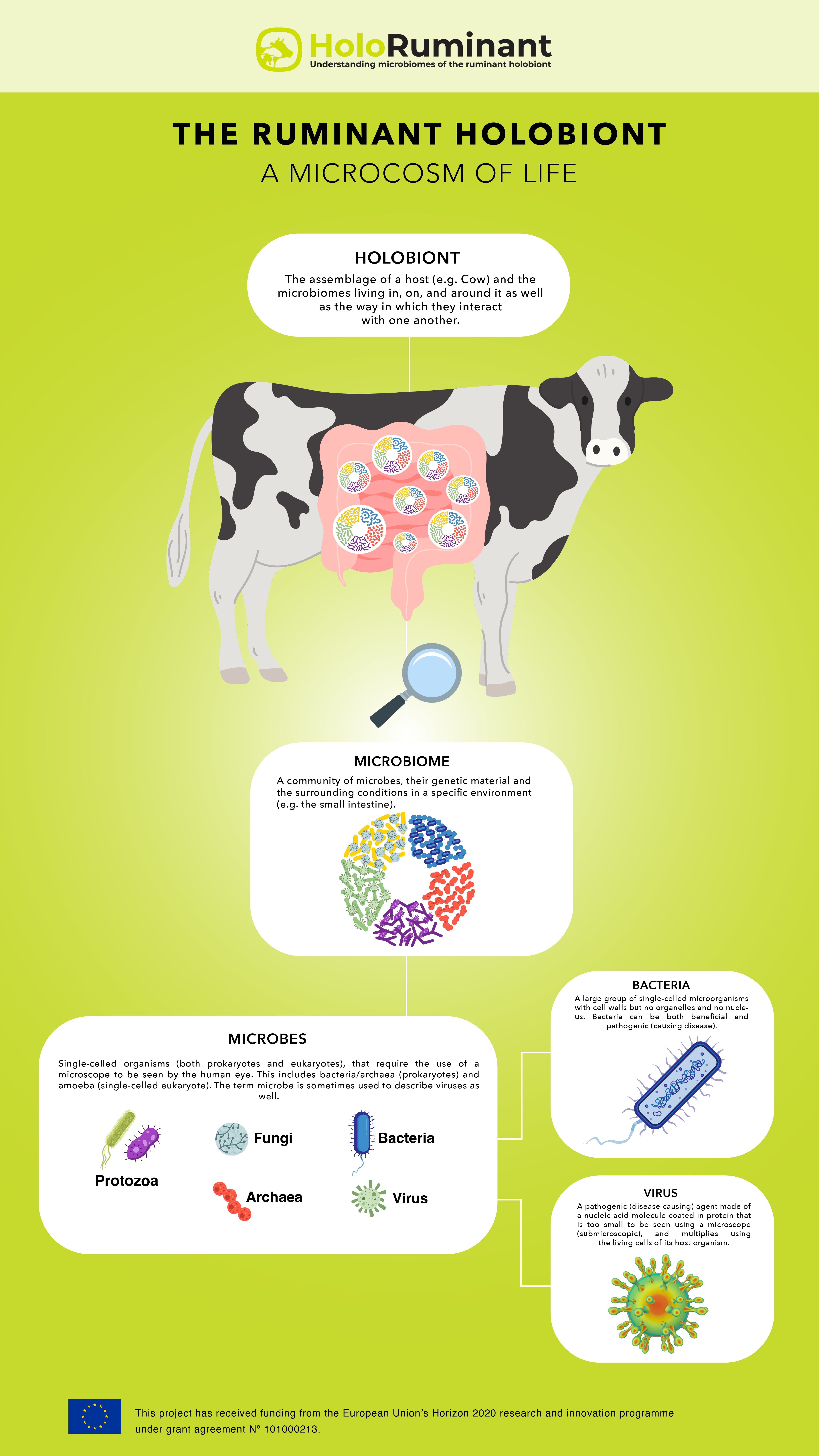
BACTERIA: A large group so single-celled microorganisms with cell walls but no organelles and no nucleus. Bacteria can be both beneficial and pathogenic (causing disease).
BAYESIAN MODEL: Mathematical formulation for learning from experience. In light of the evidence provided by data (a likelihood), pre-existing beliefs (a prior) are updated to reflect beliefs better aligned with the observed data (a posterior).
BIG DATA: Extremely large and complex datasets that are challenging to store, process, analyze, and interpret. Data scientists often need to use specialized tools and methods to work with big data.
BIOLOGICAL EFFICIENCY: Biological efficiency refers to comparing some aspect of animal performance at the individual or herd level. Biological efficiency may include traits such as energy conversion, growth and offspring production.
BROAD-SENSE HERITABILITY: The extent to which the genotypes determine individuals’ phenotypes.
CHROMATIN ACCESSIBILITY: the level of physical compaction of chromatin.
CHROMATIN: Structure made of DNA and proteins that enable the DNA to be packed in the nucleus.
CHROMOSOME: the basic structure into which the DNA is arranged inside the cells.
CODING REGIONS: The coding region of a gene are the parts of a gene’s DNA that codes for a protein.
CODING: Using a programming language to communicate with a computer and provide it with instructions, referred to as an algorithm.
COMPARATIVE GENOMICS: In comparative genomics, the genomic features of different organisms are compared. Examples of such genomic features are genes and gene order.
COMPLEX TRAIT: Complex traits do not follow the principle of simple Mendelian inheritance, where one gene codes for one trait. These traits show a continuous range of variation, and both environmental and genetic factors influence them. Complex traits are also known as quantitative traits.
DATA SCIENCE: Interdisciplinary field that combines tools from statistics, mathematics, and computer science to find interesting patterns from complex datasets, including big data.
DATASET: A structured collection of related information—numbers, measurements, words, or descriptions—that has been gathered and stored for a specific reason.
DISEASE RESISTANCE: The ability of organisms to withstand a pathogen challenge and remain virtually unaffected. Animals with higher disease resistance are therefore less likely to get sick.
DNA METHYLATION: An epigenetic mechanism that occurs by the addition of a methyl (CH3) group to DNA, thereby often modifying the function of the genes and affecting gene expression.
DNA: A long molecule that contains our unique genetic code. It holds the instructions for making all the proteins in our bodies.
EPIGENETICS: the study of changes in DNA that do not involve changes in DNA sequence.
EPIGENOMICS: The application of epigenetics to the full genome. It includes in particular DNA methylation, histone modifications, chromatin accessibility.
EXONS: The part of the gene that has the instructions to make a protein.
FUNCTIONAL ANNOTATION: The process of identifying genetic regions and adding information on function of these regions.
FUNDAMENTAL BIOLOGY: This branch of biology focuses on understanding the basic mechanisms of life and using this knowledge to understand many of the challenges in everyday life, from animal health and disease to loss of biodiversity and environmental quality.
GENE EXPRESSION: Gene expression is the process by which the instructions in our DNA are converted into a functional product, RNA.
GENE: Section within the genome that carries the information to make a molecule, usually a protein. They contain the instructions for our individual characteristics, like eye and hair colour. In humans and other complex organisms, genes are split into coding (exons) and non-coding sequences (introns). By variable use of these sections, a gene can make more than one type of protein.
cdabkbckjdbvkjbdkvbk
GENETIC MARKER: A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species.
GENETIC VARIATION: Differences in DNA between individuals that make them different.
GENETICS: Genetics is a branch of biology concerned with studying genes, genetic variation, and heredity in organisms.
GENOME BY ENVIRONMENT INTERACTION: Some traits are strongly influenced by genes, while other traits are strongly influenced by the environment. The interplay between genes and the environment is called genome by environment interaction. Most traits, however, are controlled by one or more genes interacting in complex ways with the environment.
GENOME EDITING: Intentionally altering a DNA sequence in a living cell.
GENOME: All the genetic material in the chromosomes of a particular organism.
GENOMIC FEATURE: Genomic regions with a known function.
GENOMIC PREDICTION: Computing approach that predicts the phenotypes of animals only based on their genetic information (by scoring DNA markers such as SNPs). The output of this calculation is named breeding value.
GENOMICS: Genomics is a discipline that studies the structure and function all of the genes of an organism (the genome), including the interactions of the genes with each other and the organism’s environment.
GENOTYPE: The complete set of genes possessed by an organism.
HEREDITY: The passing on of characteristics genetically from one generation to another.
HERITABILITY: Heritability is a measure of the contribution of genes to any trait in an animal population. It provides an estimation of how an animal will transmit its genetic characteristics to the next generation. Heritability can be calculated for any trait that can be measured.
HISTONE MODIFICATION: Modifications to histones that regulate the physical properties of chromatin and consequently these modifications may affect gene expression.
HISTONE: A protein found in the nucleus of eukaryotic cells, which is involved in packaging DNA into structural units called nucleosomes.
INTRONS: Part of a gene that is not used to make a protein and is cut out from the RNA between transcription and translation.
MACHINE LEARNING: The use of algorithms to teach a computer how to automatically learn from data and improve from experience without help from a human.
METAGENOMICS: The study of the pool of all the DNA in a sample belonging to various (micro)organisms, e.g. the content of a rumen or tissue.
METHANE: CH4 – gas produced by microbial processes, including in the rumen; a fuel and potent greenhouse gas. CH4- in the rumen, potent greenhouse gas produced by microbial processes.
MICROBES: Single-celled organisms (both prokaryotes and eukaryotes), that require the use of a microscope to be seen by the human eye. This includes bacteria/archaea (prokaryotes) and amoeba (single-celled eukaryote). The term microbe is sometimes used to describe viruses as well. Note: there is debate about microbe/microorganism… some use the terms interchangeably, others feel that they are not the same, in that all microbes are microorganisms but not all microorganisms are microbes….
MICROBIABILITY: In analogy to heritability, microbiability refers to the proportion of the host-phenotypic variance the microbiota explains.
MICROBIAL FERMENTATION: During the microbial fermentation process, microbes break down the food eaten by animals, such as plants, without using oxygen. This produces products like volatile fatty acids (like acetate, propionate and butyrate), microbial protein, and vitamins, which provide energy and nutrition for the animal. Microbial metabolic activity that takes substrates such as the plant material ingested by the host animal and, in the absence of oxygen, produces by-products, such as volatile fatty acids (for example, acetate, propionate, butyrate), microbial protein and vitamins, which constitute the host’s primary energy and nutrient sources.
MICROBIAL INFLUENCE: The influence of the microbes’ action/presence/abundance on a host phenotype of interest.
MICROBIAL MARKERS: Microbial genes, organisms, or by-products used as indicators of change in an animal phenotype.
MICROBIOME: Community of microbes, their genetic material and the surrounding environmental conditions in a defined source.
NARROW-SENSE HERITABILITY: The extent to which phenotypes are determined by the genes transmitted from the parents.
NUCLEUS: A structure in cells that contains the genome and acts as the control room for the cell.
OMICS: A term describing the study of genetic/protein/chemical information from an organism or group of organisms, such as microbial communities.
OPEN SOURCE: Type of computer software that is community developed and supported. Open source code and software.
PATHOGEN: Pathogens are organisms that can cause disease.
PHENOTYPE: All the observable physical characteristics of an organism, including colour, shape, size, biochemical properties and, performance which result from the expression of its genotype in a given environment.
PRECISION BREEDING: Precision breeding uses innovative techniques such as sensors and detectors and novel breeding techniques such as genomic selection and genome editing. By using these techniques, breeders can target specific traits and parts of the genome for selection, allowing faster achievement of breeding goals.
RNA: A nucleic acid similar in structure and properties to DNA, but it only has a single strand of bases. RNA is the messenger that transfers the information contained by DNA to make a protein.
RUMINANT: Herbivorous mammals, including but not limited to sheep, goats, and cattle, with a 4-chambered stomach in which nutrients from plant material are accessed via microbial fermentation.
SELECTIVE BREEDING: Choosing parents with particular characteristics to breed and produce offspring with more desirable characteristics.
SEQUENCING: The method of determining the order of letters or bases in DNA, or the order of amino acids in a protein molecule.
SIMULATION: A simplified imitation of a real-world system in a controlled environment.
SNP: a Single Nucleotide Polymorphism or SNP is a variation at a single position in a DNA sequence among individuals.
SOFTWARE PACKAGES: An organized collection of related algorithms that work together for a particular task or have a similar function.
THE CENTRAL DOGMA: The process by which the instructions in DNA are converted into a function product. The central dogma explains the flow of genetic information, from DNA to RNA, to make a functional product, a protein.
TRAITS: a distinguishing quality or characteristic.
VIRUS: A pathogenic (disease causing) agent made of a nucleic acid molecule coated in protein, that is too small so be seen using a microscope (submicroscopic), and multiplies using the living cells of its host organism.
ZOONOSES/ZOONOTIC DISEASES: Diseases transmitted from vertebrates, non-human mammals, to people.

